GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.
Python package for analysis of multiomic single cell RNA-seq and ATAC-seq. - GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells

Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells
GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.

GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.
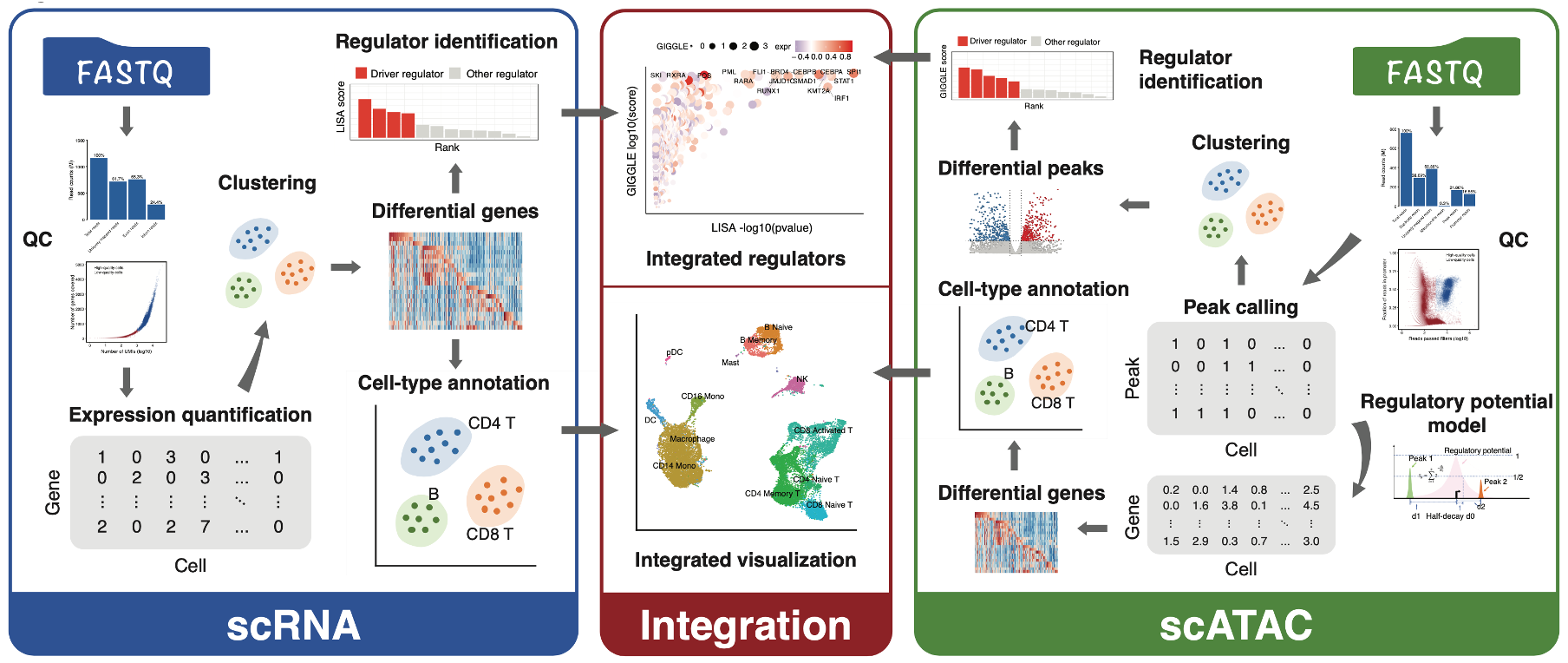
MAESTRO Single-cell Transcriptome and Regulome Analysis Pipeline
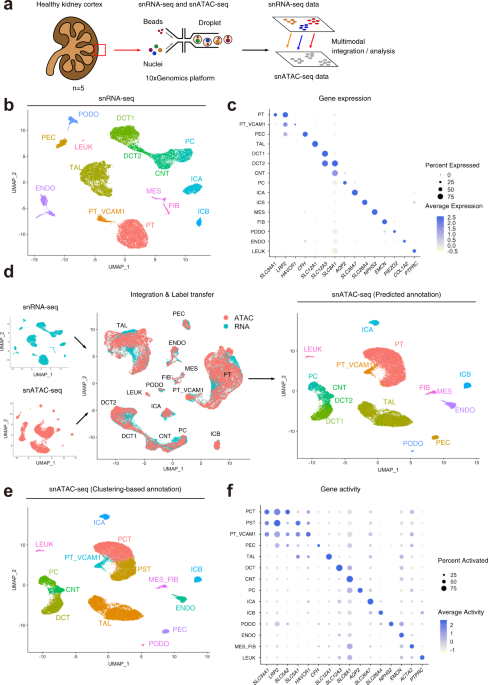
Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney
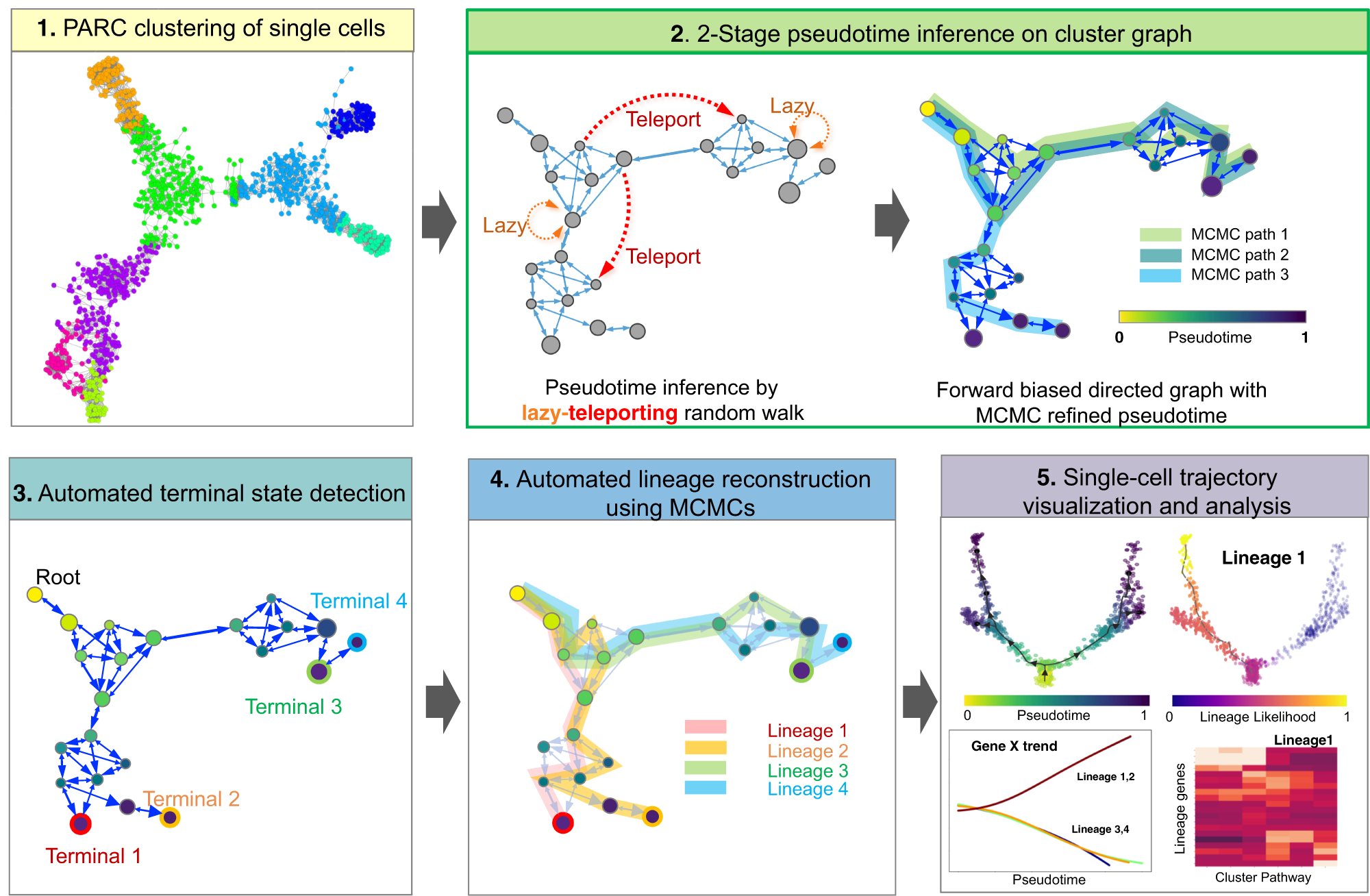
Generalized and scalable trajectory inference in single-cell omics data with VIA

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells

A unified computational framework for single-cell data integration with optimal transport









